Green Magic. A MAGIC design to study photosynthesis and nutrient homeostasis in Chlamydomonas reinhardtii
Objectives
The majority of primary producers performs oxygenic photosynthesis reducing small energy-poor inorganic compounds (CO2 and H2O) into complex energy-rich compounds (sugars and ATP) that can be sequentially used to produce new biomass. Photosynthetic organisms have colonized a wide range of habitats and developed strategies to adapt to ever-changing environmental conditions. Even within the same species, as in the case of the unicellular green alga Chlamydomonas reinhardtii, extensive genetic and phenotypic diversity can be observed. The level of variation present within a population defines the adaptative potential of the species to environmental changes. Moreover, intra-specific genetic diversity can be used as valuable source of information to understand the polygenic nature of quantitative traits and discover novel alleles responsible for phenotypes of interest.
Dissecting and understanding the genetic architecture of complex traits, such as photosynthesis, is a fundamental step toward innovation of plant breeding programs and genetic selection. Increasing demand for global food production counterposed to the rise of alarming environmental issues represents one of the biggest challenges of this century. With the shrinking of farmlands, increasing the efficiency of photosynthesis is essential to face the challenge improving crop plants productivity. Despite of the economic and ecological importance of photosynthesis, the natural genetic diversity present within and among species is still largely unexplored field.
Our project is aimed to give new insights about the complex genetic architecture of photosynthesis. To do so, we intend to take advantage of the intra-specific genetic variability naturally occurring among different strains of the photosynthetic model organism Chlamydomonas reinhardtii.
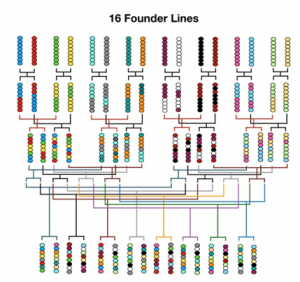
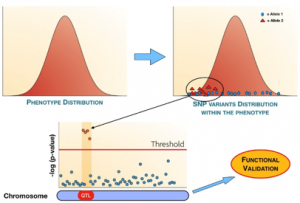
Sixteen wild type strains selected on the bases of their genotypic and phenotypic features will be inter-crossed for several generations developing a Multi-parent Advanced Generation Inter-Cross (MAGIC) design. The result of multiple generations of recombination is a population composed of about 1000 final Advanced Inter-Crossed Lines (AILs) sharing small segments of genome coming from each initial founder.
The MAGIC population will be used as a resource to develop a Genome-Wide Association Study (GWAS) mapping novel Quantitative Trait Loci (QTLs) correlated to the photosynthetic phenotype. The QTLs will be successively annotated identifying and characterizing the gene variants causing the variation of the associated phenotypic trait.
PROJECT LEADERS
Tom Druet, PhD
FRS-FNRS Senior Research Associate
GIGA / Medical Genomics-Unit of Animal Genomics

Pierre CARDOL, PhD
FRS-FNRS Senior Research Associate
Génétique et physiologie des microalgues
Marc Hanikenne, PhD
FRS-FNRS Senior Research Associate
Génomique fonctionnelle et imagerie moléculaire végétale
RESEARCHERS
Fabrizio Iacono, PhD Student
Génétique et physiologie des microalgues
Sara Esteves, PhD Student
Génomique fonctionnelle et imagerie moléculaire végétale
Alice Jadoul, Technician
Génomique fonctionnelle et imagerie moléculaire végétale